Background
A brain tumour can be defined as the uncontrolled development of cancerous cells in the brain. According to previous research, if left unchecked, a brain tumour can lead to cancer. Therefore, it is essential for a radiologist to be accurate about the existence of brain tumours from magnetic resonance images (MRI) for their analysis.
With the advent of e-health and machine/deep learning techniques, medical specialists are able to provide better health care and quick responses to their patients. Using machine learning (ML) techniques, an ML model can be trained to know if brain tumours are in MRI images. Machine learning is a branch of artificial intelligence that can by themselves learn how to solve specific problems if given the right access to data. Furthermore, ML has been effective in making decisions and predictions from data produced by healthcare industries.
This article will critically review different ML pipelines and models used in detecting brain tumours from MRI images and evaluate their strengths and limitations. The datasets used for analysis in this article are the T1-CE MRI image dataset, TCIA (The Cancer Imaging Archive), and Rembrandt database for brain cancer imaging.
Methods and analysis
In this article, a deep neural network called Convolutional neural network and two traditional machine learning algorithms called K-Nearest Neighbours and Nave Bayes’ for detecting cancer tumours in the human brain using MRI images in this study.
Method 1.
Convolutional Neural Networks (CNN):
A Convolutional Neural network is a method of deep learning that uses convolutions on a kernel that slides through an image and produces a feat map to better understand segments and objects within an image. A convolutional neural network is used here to segment brain tumour into one of various four classes:
- Healthy region
- Meningioma tumour
- Glioma tumour
- Pituitary tumour
This article will not emphasise which architecture performs best, but on some aspects that are worth taking note of when training a CNN.
A CNN’s basic structure consists of an input image, a kernel or filter (usually a 3 x3) matrix that slides horizontally across the image repeatedly moving X strides at a time and generating an output. The weights are then adjusted depending on how alike the newly generated feature map compares to the original input image. The basic structure might sound simple, but many actors come into play for the algorithm to be able to segment and locate brain tumour cells accurately. This study summarizes some of these aspects:
CNN architecture: figure 1 below shows the CNN architecture used to classify the different tumours.
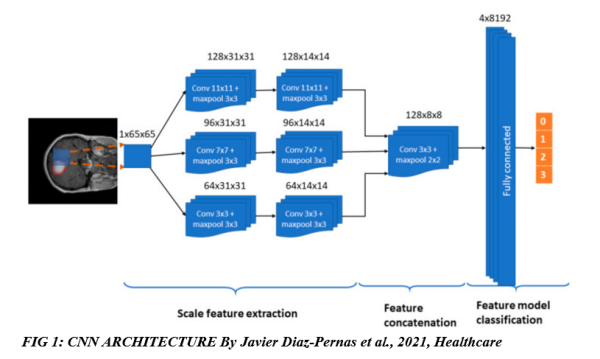
The figure above shows how the CNN architecture processes an image pixel by pixel and automatically extracts the features needed and classifies the tumours using one of four different labels from 0 to 3; 0 – healthy region, 1 – meningioma tumour, 2 – glioma tumour, and 3 – pituitary tumour.
Overfitting: This is a very important issue for CNN. When the machine learning algorithm overlearns or memorises the train data, it cannot generalise properly on unseen data. This issue can be taken care of by using more artificially generated data in data augmentation, which is a popular method for this. Another method to avoid overfitting is using dropouts, which is dropping out a certain percentage of the neurons in the network to prevent overlearning. Other methods of dealing with this issue include batch normalisation and pooling.
Batch normalisation is a method of normalisation in the data that employs mini batches, which speeds up the training process by reducing the normal of epochs to be trained and stabilising the training process.
Pooling is another important aspect that downsizes the image and causes the machine learning algorithm to learn features on a downsized or less detailed image. Different pooling methods exist, such as max pooling, which uses the maximum value from the pool to estimate, while mean pooling uses the mean as an estimator.
As the data is non-linear, they need a function to introduce non-linearity in the data. The right activation function for this is the ReLU function or the rectilinear unit. After several layers of convolutions and rectifying using the RelU, the data is completely flattened using pooling into a columnar matrix which is then passed through a fully connected layer. Using a SoftMax activation, the fully connected layers can then be classified based on the classes initiated. The feature map gotten from this will then be used to classify MRI images based on the features it has learned. Keras API was used here as it is a framework for object detection and segmentation.
Method 2
K-Nearest Neighbours (KNN):
K-Nearest Neighbours (KNN) is a classical shallow machine learning algorithm used for brain tumour segmentation and classification. In this study, MRI images undergo segmentation via k-means clustering, an unsupervised algorithm. Features extracted from these clusters are then analyzed using the Gray level Co-Occurrence matrix (GLCM) and inputted into the KNN classifier for classification.
KNN requires extensive data pre-processing to achieve significant results. The study focuses on key pre-processing techniques, including image enhancement through filtering and resizing. Filtering techniques such as mean and median filters are employed to eliminate noise like salt and pepper, Gaussian noise, speckle, and Brownian noise.
Image segmentation involves creating clusters based on color, texture, contrast, and brightness. Cluster analysis using the unsupervised algorithm k-means facilitates easy feature extraction.
Feature extraction utilizes the Gray level Co-occurrence matrix, which measures the spatial dependence of grey-level intensities between pixels. This method has shown accurate results (89.9%) in classifying brain tumour cells using MRI images.
Once features are extracted, they are fed into the KNN classifier, with each segment representing a distinct class. The focus of this article is not on the specific configurations or steps taken by KNN for classification of the feature set.
Method 3
Naïve Bayes:
The Naïve Bayes algorithm is a supervised machine learning technique used for classification based on the probabilistic theory of Bayes. It assumes that all features (pixels) are independent of each other, making it suitable for applications with randomness.
Similar to the KNN method discussed earlier, the Naïve Bayes algorithm requires important pre-processing steps to prepare the data for the machine learning process. The accuracy of the model heavily relies on these pre-processing steps. This article focuses on the following pre-processing techniques:
- Image Enhancement: One crucial technique is noise removal from the original MRI image. Salt and pepper noise can be eliminated using a median filter. The image is converted to grayscale and resized to ensure consistent sizes throughout the dataset. Additionally, a method called pixel extraction is applied to extract the unwanted region (skull) from the original MRI image.
- Image Segmentation: Segmentation involves distinguishing abnormal issues from normal brain tissues. Various techniques such as regional growing, edge-based, clustering (as discussed in the KNN method), watershed, deformable model-based, and threshold technique can be employed. The threshold method relies on predefined thresholds for image segmentation.
- Feature Extraction: This study utilizes the Discrete Cosine Transform (DCT) for feature extraction. DCT decorrelates the image data into 2-dimensional input data, enabling effective feature representation.
By implementing these pre-processing techniques, the Naïve Bayes algorithm can be applied for accurate brain tumour classification.
After all the above pre-processing steps, the data is now ready to be fed into the Naïve Bayes classification algorithm, whose configuration shall not be discussed in this article.
Discussion & evaluation
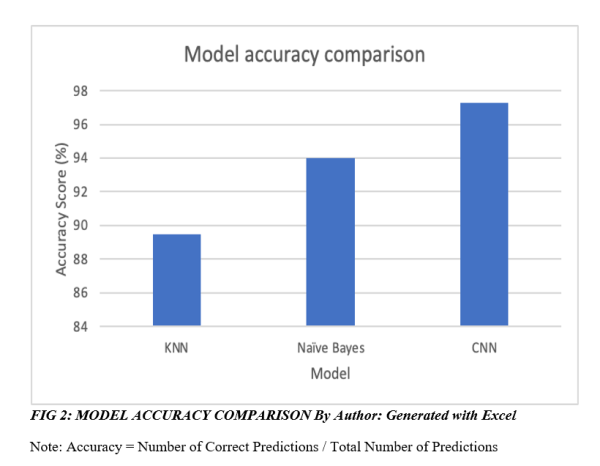
When looking at the results from the three different methods, it is clearly seen that the use of machine learning in detecting a brain tumour from MRI scans is very promising, with all three methods producing a high level of accuracy. In the classification process, model validation is used to divide the data into training and testing in order to obtain the accuracy of testing results.
The result of the three models in this study can be seen in the figure below. However, all three models were trained and tested on different datasets, which should be noted when comparing accuracy.
Final thoughts
As machine learning gains traction in technology and e-health industries, it’s vital to recognize how different models and pipelines impact performance.
The deep learning model Convolutional Neural Network (CNN) outperformed K-Nearest Neighbour Network (KNN) and Naïve Bayes models in this study, despite lacking spatial information and potential pooling issues. However, performance comparison was based on three distinct datasets, limiting accurate assessment.
When selecting a model, dataset characteristics like size and complexity are crucial. Deep learning models excel in large datasets with intricate patterns, but require powerful GPUs. In contrast, traditional machine learning models thrive with smaller data volumes.
https://uk.linkedin.com/in/solomonegwu
(written by Solomon Egwu)
Media Contact
Company Name: Solomon Chidi Egwu
Contact Person: Solomon Chidi Egwu
Email: Send Email
Country: United Kingdom
Website: https://github.com/solomonsquare
